WE USE MULTIDISCIPLINARY APPROACHES
Our laboratory uses multidisciplinary approaches to elucidate the function and regulation of RNA-binding proteins. We are using genetic, biochemical, functional genomic, and proteomic approaches as well as collaborating with structural biologists and computational biologists to achieve these goals. Notably, we take advantage of the complementarity of fission yeast and human cellular model systems to emphasize on evolutionarily conserved functions of RNA-binding proteins. We have a broad research program that can be grouped in the following projects:
- Polyadenylation-dependent gene regulation
- Extra-ribosomal Functions of Ribosomal Proteins
- Functional Coupling between transcription and RNA processing
- Nuclear RNA surveillance
BROAD RESEARCH PROGRAM
DETAILS ON FALLOWING PROJECTS
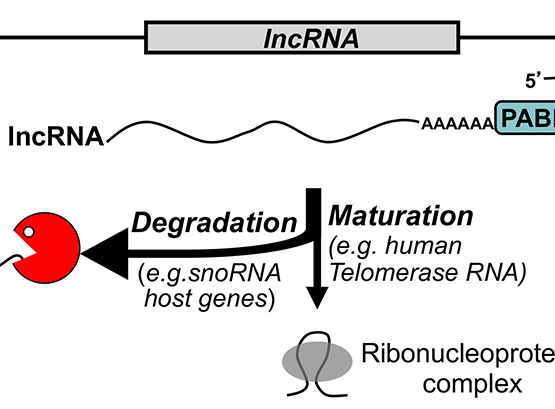
Polyadenylation-dependent Gene Regulation
Oculopharyngeal muscular dystrophy (OPMD) is an inherited disease caused by mutations in the gene encoding PABPN1 (Poly(A)-Binding Protein Nuclear 1).
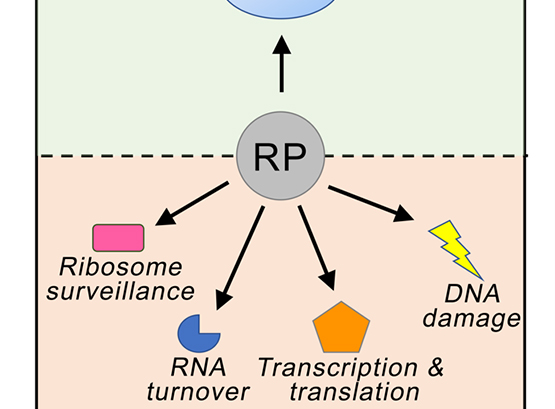
Extra-ribosomal functions of Ribosomal Proteins
The general view of ribosomal proteins (RPs) is one of generic cellular proteins that function exclusively to maintain the integrity of the ribosome.
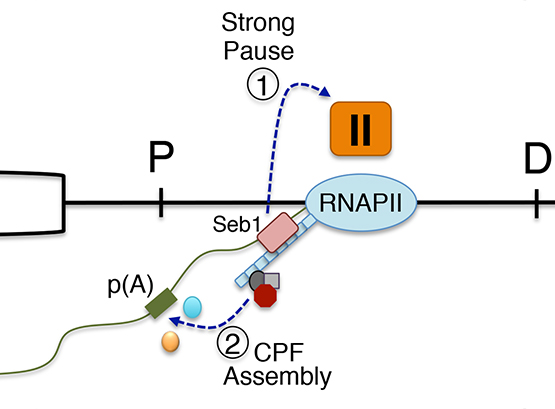
Functional Coupling between Transcription and RNA Processing
We are highly interested in understanding how various steps of RNA processing are intimately coupled to transcription by RNA polymerases.
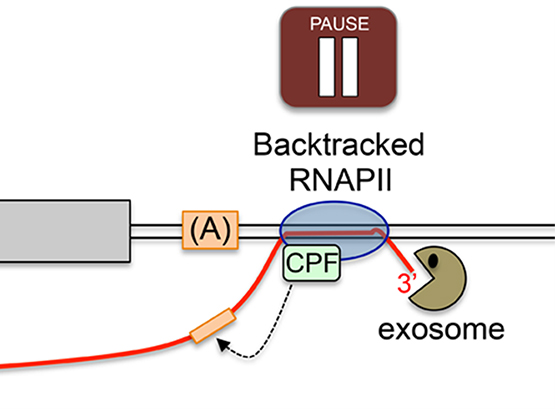
Nuclear RNA surveillance
Recent studies indicate that transcription is pervasive in eukaryotes, producing a complex network of noncoding RNAs (ncRNAs) that exist stably in cells or are rapidly degraded.